Utilizing artificial intelligence, UT Southwestern scientists have identified a new loved ones of sensing genes in enteric microbes that are joined by framework and almost certainly purpose, but not genetic sequence. The findings, published in PNAS, provide a new way of determining the purpose of genes in unrelated species and could direct to new means to struggle intestinal bacterial bacterial infections.
“We recognized similarities in these proteins in reverse of how it truly is normally completed. Alternatively of making use of sequence, Lisa seemed for matches in their framework,” reported Kim Orth, Ph.D., Professor of Molecular Biology and Biochemistry, who co-led the examine with Lisa Kinch, Ph.D., a bioinformatics professional in the Office of Molecular Biology.
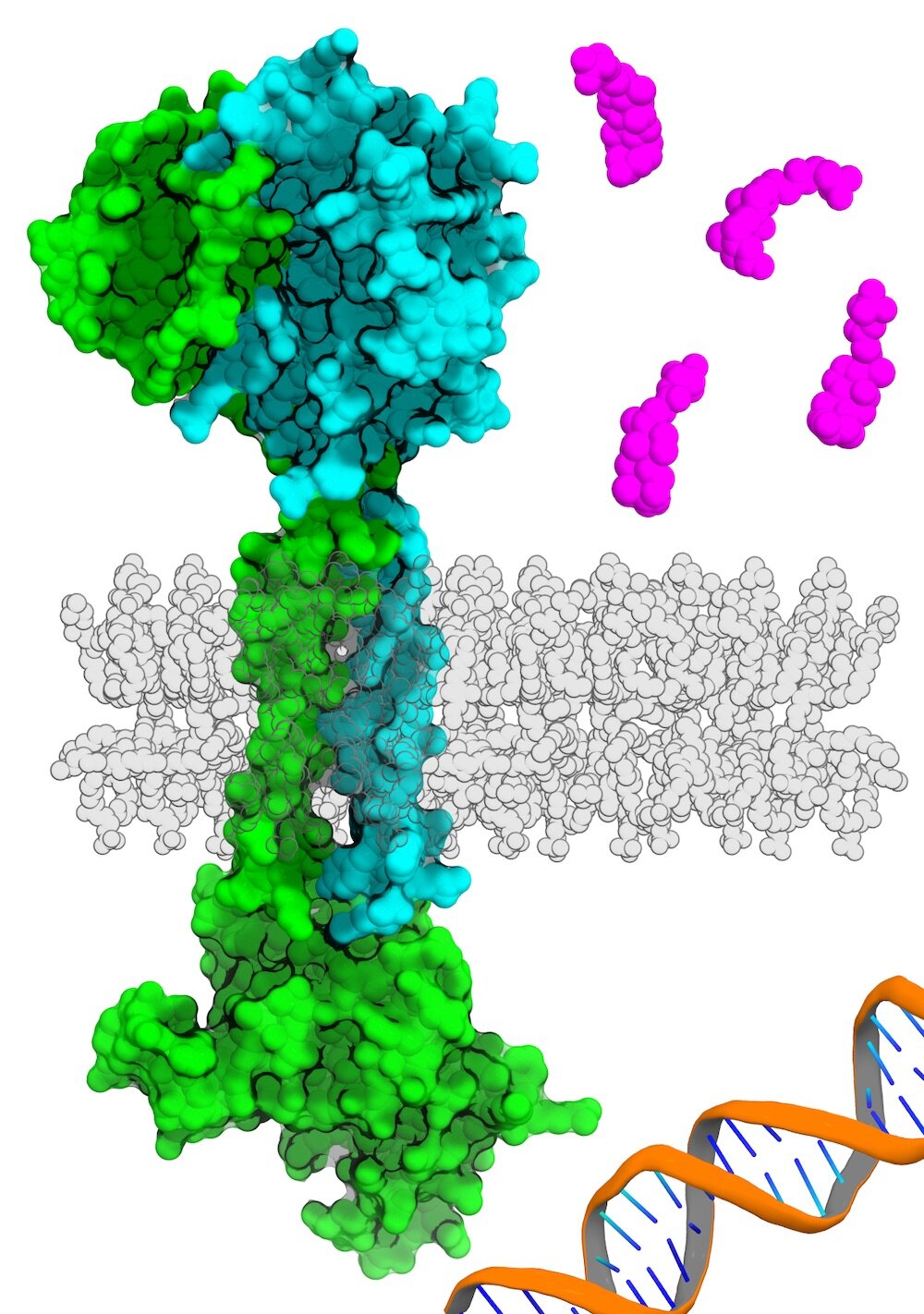
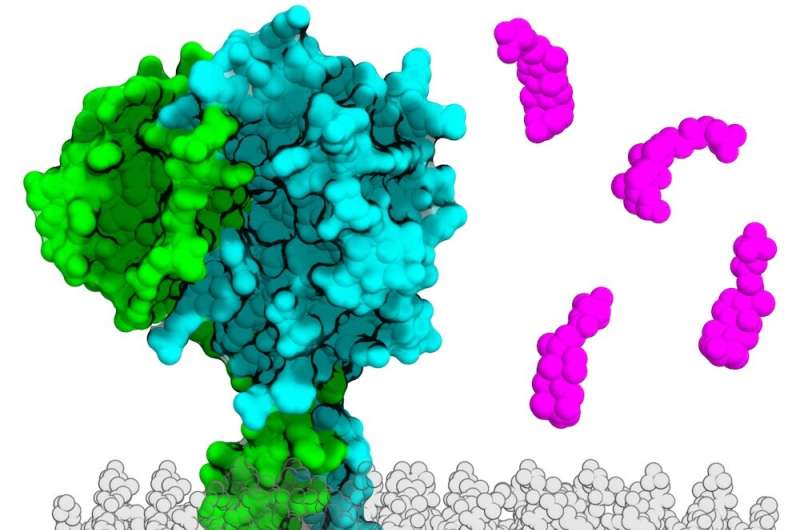
Dr. Orth’s lab has prolonged concentrated on finding out how maritime and estuary micro organism induce bacterial infections. In 2016, Dr. Orth and her colleagues applied biophysics to characterize the structure of two proteins identified as VtrA and VtrC complex that function in concert in a bacterial species recognized as Vibrio parahaemolyticus. She and her crew then found out the VtrA/VtrC sophisticated in V. parahaemolyticus—which is often the trigger of foodstuff poisoning from contaminated shellfish—senses bile from the bacterial mobile surface area, sending a signal to launch a chemical cascade that prompts this microbe to invade the intestinal cells of its human host.
Though VtrA shares some structural functions with a protein named ToxR located in a associated germs called Vibrio cholerae that causes cholera, it was unclear no matter whether a homolog for VtrC also existed in this or any other microbes.
“We had by no means found anything like VtrC,” claimed Dr. Kinch. “But, we thought, other proteins like it should exist.”
Without the need of any acknowledged genes with sequence identities equivalent to VtrC, the scientists turned to software released just two decades ago identified as AlphaFold. This synthetic intelligence program can correctly predict the composition of some proteins primarily based on the genetic sequence that codes for them—information that earlier was only gleaned as a result of laborious function in the laboratory.
AlphaFold confirmed that a protein termed ToxS in V. cholerae is quite identical in composition to VtrC, even while the two proteins did not share any recognizable parts of their genetic sequences. When the scientists searched for proteins with comparable structural characteristics in other organisms, they observed homologs for VtrC in many other enteric microorganisms species dependable for human condition, including Yersinia pestis (which causes the bubonic plague) and Burkholderia pseudomallei (which causes a tropical an infection known as melioidosis). Each of these VtrC homologs seems to do the job in live performance with proteins structurally comparable to VtrA, suggesting that their roles could be the identical as these in V. parahaemolyticus.
Dr. Orth claimed these structural similarities could sooner or later lead to prescription drugs that take care of problems brought about by distinct infectious organisms that rely on comparable pathogenic tactics.
Analyze finds sensing system in foods poisoning bug
Lisa N. Kinch et al, Co-component sign transduction units: Quick-evolving virulence regulation cassettes discovered in enteric germs, Proceedings of the National Academy of Sciences (2022). DOI: 10.1073/pnas.2203176119
UT Southwestern Clinical Centre
Quotation:
Scientists use AI to detect new family of genes in gut microbes (2022, July 2)
retrieved 2 July 2022
from https://phys.org/information/2022-07-ai-household-genes-gut-micro organism.html
This doc is issue to copyright. Aside from any fair working for the reason of personal examine or investigate, no
element might be reproduced with no the published authorization. The information is provided for data applications only.